金桔
金币
威望
贡献
回帖0
精华
在线时间 小时
|
登陆有奖并可浏览互动!
您需要 登录 才可以下载或查看,没有账号?立即注册


×
来源:知乎
来源:IVD技术咖
CRISPR的每一次登场,总是自带话题性,从基因编辑,到专利之争,再到下一代分子诊断技术,特别是在分子诊断行业,总流传着它的传说。
一、CRISPR/Cas系统简介
CRISPR的全称是Clustered Regularly Interspaced Short Palindromic Repeats(成簇规律间隔的短回文重复序列),形如其名,CRISPR 序列结构正是成簇的规律间隔的短回文重复序列,该序列与CRISPR associated(Cas)蛋白组成CRISPR/Cas系统。CRISPR/Cas系统是原核生物的一种自我保护机制,是细菌用来抵抗外来遗传物质入侵的适应性免疫防御系统。其发挥免疫的功能包括适应、表达和干扰3个阶段。在适应阶段,细菌通过Cas蛋白(Cas1和Cas2)捕捉入侵的外来核酸,并整合至自身基因组的CRISPR 序列中形成记忆功能,以便下次出现入侵时,细菌可启动免疫反应;在表达阶段,CRISPR 序列转录生成前体crRNA (pre-crRNA)和反式激活RNA(trans-activating crRNA, tracrRNA),pre-crRNA通过加工,形成含有与外来基因序列匹配的crRNA,crRNA与tracrRNA通过碱基配对形成向导RNA(single-guide RNA,sgRNA),用于Cas蛋白的募集;在干扰阶段,sgRNA引导Cas蛋白定位到与其同源的外来核酸靶标,并激活Cas蛋白的内切酶活性,对靶标核酸进行切割,从而发挥免疫功能。
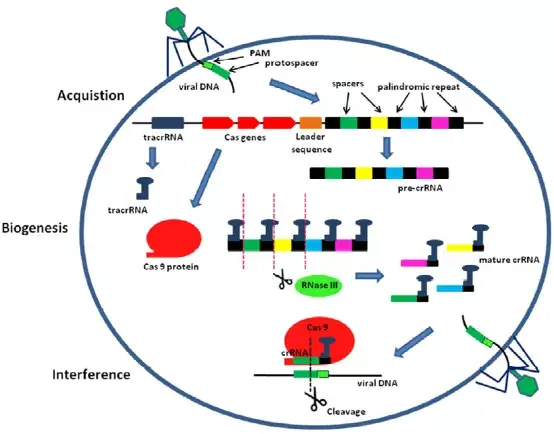
二、CRISPR/Cas分子诊断技术的建立
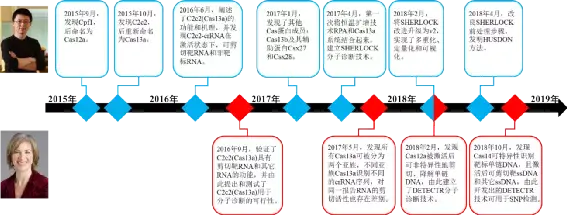
提起CRISPR,我们就不能不说起麻省理工的张锋和加州大学伯克利分校的Jennifer Doudna。两者不但在CRISPR基因编辑上做出奠基性贡献,也对CRISPR在分子诊断行业的应用做了开拓性工作。正是两位大牛在2015~2018年这四年的你来我往中,开启了CRISPR分子诊断这一全新领域。下面,我们来看看高手之间的过招,是如何让吃瓜群众拍案叫绝的。
(1)2015年9月,张锋团队在Cell发表论文,报道了CRISPR/Cas系统中另一种RNA向导的核酸内切酶(Cpf1),后改名为Cas12a,日后其将在CRISPR/Cas分子诊断系统发挥重要作用。
(2)2015年10月,张锋团队在Molecular Cell发表论文,通过在基因库里“钓鱼”,又发现了三个新的CRISPR/Cas系统效应蛋白,分别为C2c1, C2c2, 和C2c3,其中C2c2蛋白后来被重新命名为Cas13a,并成为开启CRISPR分子诊断大门的第一把钥匙。
(3)2016年6月,张锋团队在Science发表论文,进一步阐述了C2c2(Cas13a)的功能和机理,与Cas9不同,C2c2(Cas13a)仅需在crRNA 的引导下,即可作用于入侵病原体的单链RNA;更为重要的是,C2c2-crRNA一旦与匹配的RNA结合,进入激活状态后,除了特异性剪切靶RNA外,还启动了非特异性内切酶活性,即对任何单链RNA进行剪切;这就开始显现出了CRISPR/Cas系统作为分子诊断工具的特性,但该论文当时并未将C2c2(Cas13a)的特性和诊断技术联系起来。
(4)2016年9月,Doudna团队在Nature发表论文,研究了来源于不同菌类的C2c2(Cas13a),发现C2c2(Cas13a)还有将pre-crRNA加工成crRNA的功能,同时验证了张锋团队关于C2c2(Cas13a)具有特异性剪切靶RNA和非特异性降解其它RNA功能的结论,并由此提出和测试了C2c2(Cas13a)用于分子诊断的可行性。
(5)2017年1月,张锋团队在MolecularCell发表论文,再次通过“钓鱼”方法发现了其他Cas蛋白成员:Cas13b及其辅助蛋白Csx27和Csx28,其中Cas13b为以后实现多重检测带来了可能性。
(6)2017年4月,张锋团队在Science发表论文,第一次系统介绍了CRISPR/Cas作为分子诊断技术的应用,该方法将恒温扩增技术RPA和Cas13a系统结合起来,实现了对病原体高特异性、高灵敏度的检测,命名为Specific High Sensitivity Enzymatic Reporter UnLOCKing,简称SHERLOCK。
(7)2017年5月,Doudna团队在Molecular Cell发表论文,发现所有Cas13a可被分为两个亚族,不同亚族Cas13a识别不同的crRNA序列,激活后的Cas13a-crRNA对同一报告RNA的非特异剪切活性也存在差别,这为CRISPR/Cas系统实现多重检测建立了理论基础。
(8)2018年2月,Doudna团队和张锋团队分别在Science发表了一篇论文。Doudna团队主要发现Cas12a被激活后可非特异性地剪切、降解单链DNA,并以此建立了另一个CRISPR/Cas分子诊断技术,命名为DNA Endonuclease-targeted CRISPR Trans Reporter,简称DETECTR,为了提高灵敏度,该方法同样引进了RPA对靶标进行扩增放大产物。张锋团队的同期论文则主要对SHERLOCK进行改进升级,实现了该方法的多重化、定量化和可视化,命名为SHERLOCK v2,为了实现多重检测,他们通过筛选确定了不同来源的Cas12a、Cas13a和两个Cas13b,并基于四个Cas蛋白对应的crRNA不同,以及激活后剪切的核苷酸对差异,由此构建出了一个可实现四重检测的分析方法;为了实现定量检测,他们通过对RPA扩增阶段的引物浓度进行系列稀释测试,筛选出合适的引物浓度,使得RPA扩增过程处于线性增长阶段,从而实现SHERLOCK在较宽样品浓度范围内的定量;最后的可视化,主要是为了实现CRISPR/Cas诊断系统在POCT的应用,他们将CRISPR/Cas检测技术与侧向层析相结合,通过肉眼可见的显色情况来判读结果。
(9)2018年4月,张锋团队继续在Science发表论文,对SHERLOCK的前处理方法进行改良,发明了Heating Unextracted Diagnostic Samples to Obliterate Nucleases,简称HUDSON,该方法不经核酸提取纯化,仅需对临床样本进行核酸酶灭活和加热等快速处理,即可用于后续SHERLOCK反应,利用HUDSON+SHERLOCK技术,在2小时内便可肉眼观测到寨卡病毒和登革热病毒的检测及分型结果。
(10)2018年10月,Doudna团队在Science发表论文,报道了另一个新的Cas蛋白——Cas14,该蛋白分子量更小,识别的靶标为单链DNA,激活后除了剪切靶ssDNA外,还非特异性地剪切其它ssDNA,而且该蛋白不受PAM序列的限制,对靶ssDNA的特异性识别能力更强,由此开发出的Cas14-DETECTR可用于SNP检测。
(11)至此,CRISPR/Cas分子诊断技术中最常用的三个Cas蛋白已悉数亮相。实际上,对于CRISPR/Cas分子诊断技术的推进发展,并非只有张锋和Jennifer Doudna两个团队在忙活,国内也有其他研究者致力于优化和改进CRISPR/Cas分子检测系统。例如:
①2018年4月,中国科学院上海生命科学研究院王金团队在Cell Discovery发表了基于Cas12a的分子诊断技术,该技术名称为one-HOur Low-cost Multipurpose highly Efficient System,简称HOLMES,该方法利用PCR对靶标进行扩增后,再利用CRISPR/Cas12a系统进行检测,后将Cas蛋白换为嗜热的Cas12b,并与恒温扩增技术LAMP(LAMP扩增原理详见聊聊恒温扩增技术-LAMP & CPA)结合,升级为HOLMES v2;
②2018年9月,中国科学院深圳先进技术研究院喻学锋团队在Nature Communications发表了基于Cas9切口酶活性开发的分子诊断技术,该技术名称为CRISPR-Cas9-triggered nicking endonuclease mediated Strand Displacement Amplification method,简称CRISDA,该方法使用等温扩增扩增技术SDA作为扩增手段,利用HNH结构域失活的Cas9切割打开靶DNA 的一条链,再用引物启动SDA 反应,最后使用肽核酸探针进行检测,该方法在复杂的背景干扰下仍具有高特异性和灵敏度。
三、CRISPR/Cas分子诊断技术的原理
1.基于CRISPR/Cas9系统特异性识别和切割DNA序列的功能
Cas9的主要应用是在基因编辑,其形成的复合物在sgRNA的引导下,通过识别靶标DNA上的PAM(protospacer-adjacent motif),特异性地与靶标DNA结合,并切割靶标DNA。基于CRISPR/Cas9系统特异性识别和切割DNA序列的功能,其分子检测方法可分为2类。
(1)基于切割功能的检测方法
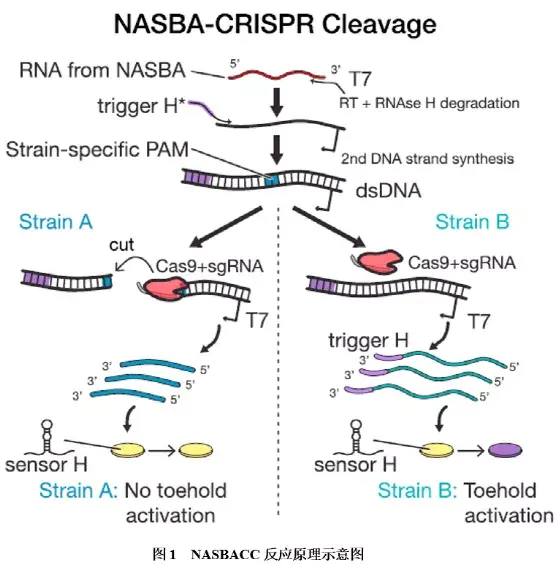
①NASBACC技术:NASBA-CRISPR Cleavage(NASBACC)技术将恒温扩增技术NASBA、toehold开关RNA传感器和CRISPR/Cas9系统进行结合,实现了寨卡病毒的分型检测,其反应原理如图1所示:首先靶标RNA通过NASBA(NASBA扩增原理详见转录介导的核酸扩增技术小结)技术在引物的设计上,引入T7转录启动子和带有toehold开关触发的序列,扩增过程中产生的双链DNA若含有特异性PAM序列和gRNA识别位点时,将被Cas9切割成两段,含有T7转录启动子的序列经T7 RNA聚合酶转录出截短的RNA,该短RNA由于缺乏传感器H触发序列,因此无法激活传感器H的toehold开关;若双链DNA不含PAM序列,则可被T7 RNA聚合酶转录出含有传感器H触发序列的全长RNA产物,从而激活传感器H;传感器H在被激活前,通过隔离核糖体结合位点(RBS)和起始密码子顺式阻断LacZ基因的翻译;当toehold开关与互补的触发RNA结合后,RBS和起始密码子被释放,激活LacZ基因翻译表达β-半乳糖苷酶,该酶可将黄色底物(氯酚红-β-D-吡喃半乳糖苷)转化为紫色产物(氯酚红),从而实现肉眼可见的颜色变化。
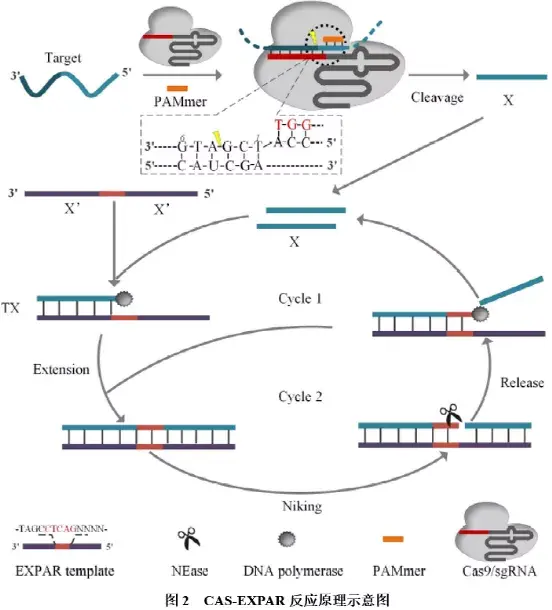
②CAS-EXPAR技术:CRISPR/Cas9 triggered isothermal exponential amplification reaction(CAS-EXPAR)是基于CRISPR/Cas9的切割反应和切口内切酶(NEase)介导的核酸扩增反应,其反应原理如图2所示:首先,Cas9与sgRNA结合形成复合物,并在PAMmer的协助下识别靶标序列,在靶标序列的特异位点切割生成目标片段X;该目标片段X可作为引物,与扩增模板结合(该扩增模板含有两个与目标片段X互补的区域X’,被NEase识别序列间隔开),并在DNA聚合酶作用下,合成含有NEase识别位点的完整双链DNA;NEase通过识别酶切位点在双链DNA上引入切口,DNA聚合酶在切口处再次延伸形成双链DNA,并置换出目标片段;置换出的目标片段可作为引物,与另一个扩增模板结合启动DNA合成,从而形成循环扩增过程,生成大量双链DNA产物,可用SybrGreen I等荧光染料进行检测。
(2)基于定位功能的检测方法
Cas9蛋白含有 HNH和 RuvC两个核酸酶结构域,用于切断dsDNA,将这两个结构域的核酸酶活性区域突变,使 Cas9失去剪切dsDNA 能力,则形成Cas9突变体dCas9。dCas9 保留了特异性靶向dsDNA的功能,通过在dCas9上融合其他功能蛋白,可构建CRISPR/dCas9检测系统。
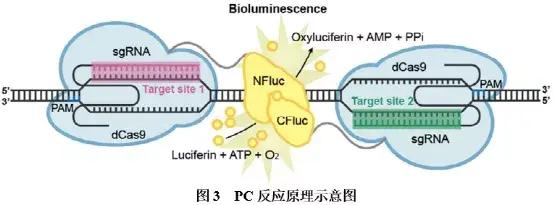
①PC技术:paired dCas9(PC)使用一对dCas9蛋白,分别融合荧光素酶的两个分离结构域(NFluc和CFluc),并通过两个靶向相邻序列的gRNA实现检测。其反应原理如图3所示:当存在靶标DNA时,两个分别带有荧光素酶结构域的dCas9,在gRNA的引导下结合到相邻的靶标序列上,荧光素酶的两个分离结构域相互靠近,形成有活性的荧光素酶,在ATP和氧气存在条件下,催化荧光素氧化,生成氧化荧光素,并产生可检测的荧光信号。
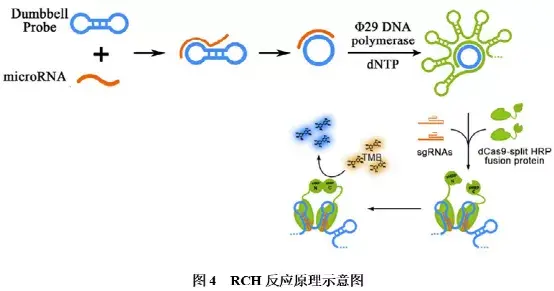
②RCH技术:RCA-CRISPR-split-HRP(RCH)结合了滚环扩增(Rolling Circle Amplification,RCA)、CRISPR/dCas9和辣根过氧化物酶(HRP)技术,可经显色反应区分miRNA的单碱基差异。其反应原理如图4所示:首先,靶标miRNA与哑铃状特异性探针结合,打开探针的双链茎部区域,形成环状探针,并在phi29 DNA聚合酶作用下进行滚环扩增,形成很多重复连续的单链DNA序列,且由于序列本身存在互补配对区域,可经互补配对形成大量规则的茎-环结构;然后,加入融合了split-HRP报告片段的dCas蛋白,并在sgRNA 的引导下,定位至扩增产物的茎-环结构,分离的HRP报告片段因此聚集,形成有活性的HRP蛋白;HRP蛋白发挥催化活性,使得黄色底物TMB转化为蓝色产物。
2.基于CRISPR/Cas系统反式切割活性的检测技术
该检测技术所用的Cas蛋白主要有Cas12、Cas13和Cas14,这类蛋白除了与Cas9一样,具有特异性切割靶序列的功能外,在激活状态下,还具有非特异性切割其它核酸序列的功能。在没有激活的状态下,Cas蛋白保持沉默,不会胡作非为;一旦被激活,便是一个没有感情的收割机,高举镰刀,看到韭菜就要割,被割下的一茬茬绿油油韭菜,即是可用来检测的信号。基于Cas蛋白的这个特点,张锋和Doudna各自创立了对应的体外诊断公司,专注于开发与CRISPR/Cas系统相关的体外诊断技术及产品。
(1)基于Cas12开发的诊断技术
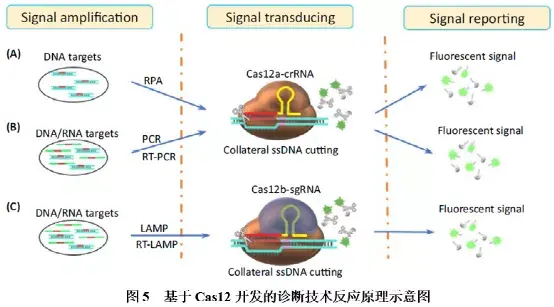
Cas12仅需在crRNA引导下即可识别靶标dsDNA,并靶向富含T的PAM位点,对靶序列进行特异性切割,同时附带非特异性切割ssDNA的活性。基于Cas12开发的分子诊断技术主要有DETECTR(图5A)和HOLMES(图5B),其中HOLMES将Cas12a更换为Cas12b后,进一步升级为HOLMES v2(图5C)。该三种方法原理类似,主要区别是对靶标扩增方法的选择不一样。首先,靶标核酸经等温(RPA或LAMP)或变温(PCR)方法扩增富集;然后,扩增产物与Cas12-crRNA结合,激活Cas12的附属切割功能,对ssDNA荧光探针进行切割,释放荧光基团,形成检测信号。
(2)基于Cas13开发的诊断技术
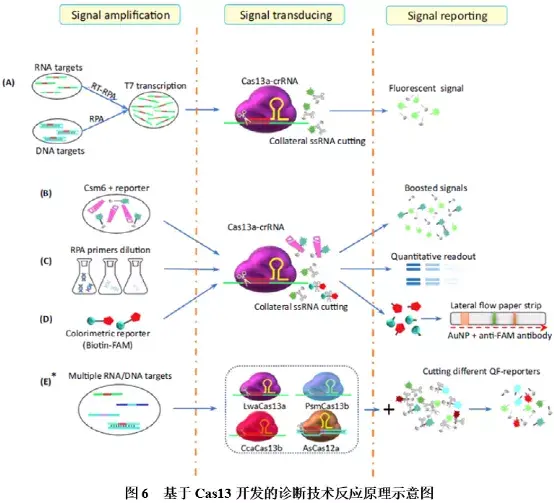
Cas13是一种RNA引导的RNA核酸内切酶,可在crRNA的引导下特异性切割单链靶标RNA,并在完成切割后,可继续保持活性并切割其他非靶标RNA。基于Cas13开发的分子诊断技术主要为SHERLOCK,其反应原理如图6A所示:靶标核酸经等温扩增技术RPA扩增并引入T7启动子序列后,再利用T7 RNA聚合酶转录形成ssRNA,转录产物与Cas13a-crRNA结合,激活Cas13a核酸酶活性,切割ssRNA荧光探针,释放可被检测的荧光基团。SHERLOCK通过进一步优化,升级成了SHERLOCK v2(图6B~E),可进行高灵敏度的多重、定量检测,结合侧向层析技术还可实现可视化检测。
(3)基于Cas14开发的诊断技术
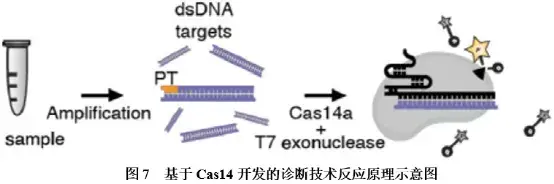
Cas14是一种RNA 引导的靶向ssDNA核酸酶,该酶分子量更小,对ssDNA的特异性识别能力更强,且不受PAM序列的限制,激活后可附带非特异性切割ssDNA的活性。基于该酶特点,Doudna团队将DETECTR技术使用的Cas12a更换为Cas14a,开发出了Cas14-DETECTR诊断技术,其反应原理如图7所示:首先,用于扩增富集靶标核酸的其中1条引物需进行硫代磷酸化(phosphorothioate, PT)修饰,由此扩增产生的dsDNA经T7核酸外切酶作用,降解其中一条未被PT保护的DNA链,从而形成ssDNA,并作为靶标ssDNA激活Cas14a内切酶活性,非特异性切割ssDNA荧光探针,生成信号。
四、CRISPR/Cas分子诊断技术的优势与不足
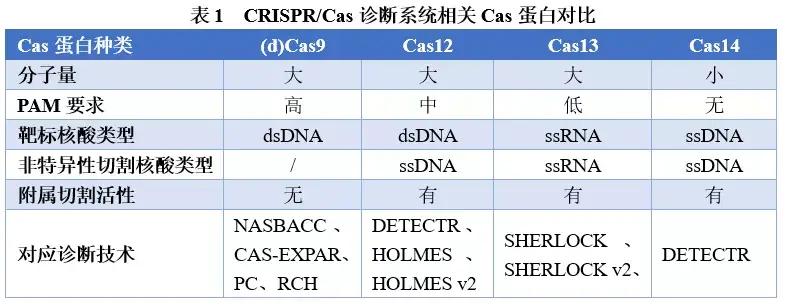
CRISPR/Cas系统作为分子诊断技术,主要是基于该系统特异性识别和切割核酸序列的功能建立的,其中切割功能中的反式切割属性,是CRISPR/Cas系统发挥体外诊断能力的最主要功能。具有反式切割活性的三种Cas蛋白(Cas12、Cas13和Cas14)被激活后,可对非靶标核酸进行切割,将检测信号进一步放大,提高了检测灵敏度,同时gRNA对靶序列的特异性识别,进一步提高了检测特异性,使得CRISPR/Cas诊断系统具有高灵敏度、高特异性的特点。不同特性Cas蛋白的发现(表1),丰富了检测核酸类型的选择,从dsDNA到ssDNA再到RNA,均可作为CRISPR/Cas分子诊断系统的靶序列,结合层析技术等显色反应,还可实现检测结果的可视化,为分子POCT领域的应用场景带来了无限可能。
CRISPR/Cas系统作为一种可特异性识别靶序列,并具有催化放大信号功能的检测技术,可直接对靶标序列进行检测,但目前的检测方法,均需预先扩增靶标核酸后,其检测灵敏度方可满足诊断要求,这就增加了操作步骤,耗时也更长了。CRISPR/Cas系统的反式切割活性具有随意性,对实现定量和多重检测的难度较大。现有Cas蛋白大部分依赖于PAM序列发挥作用,对靶标序列的选择受到限制。作为一种基因编辑技术,其脱靶效应的存在也会影响到诊断结果的准确性。但是,CRISPR/Cas系统作为下一代诊断技术的引领者,相信国内外会有研究者正在潜心改进这个技术,使得CRISPR/Cas系统更趋近于理想的诊断技术。
五、关于CRISPR/Cas分子诊断技术的一点思考
CRISPR/Cas诊断系统自发明以来,一直被大家所关注,但是目前该技术更多处于实验室测试阶段,临床上的应用价值还没展现出来。在新型冠状病毒的检测上,从张锋公布了CRISPR/Cas检测新冠的流程,到国内第一个使用CRISPR方法检测新冠产品获批,本以为该技术会从此腾飞,但还是在喧闹中慢慢沉寂了。毕竟,相比其它分子诊断技术,CRISPR/Cas分子检测系统,从发明到现在仅有几年时间,作为一种新的方法,为市场所接受还需时日。另外,作为一种专利主要集中在少数几个企业的新技术,其它企业如果推广该技术的应用,是否存在侵权风险,这也可能是大家所忧虑的。
参考资料[1] Carter J, Wiedenheft B. SnapShot: CRISPR-RNA-guided adaptive immune systems. Cell. 2015 Sep 24;163(1):260-260.[2] Hryhorowicz M, Lipiński D, Zeyland J, et al. CRISPR/Cas9 Immune System as a Tool for Genome Engineering. Arch Immunol Ther Exp (Warsz). 2017 Jun;65(3):233-240.[3] Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015 Oct 22;163(3):759-71.[4] Shmakov S, Abudayyeh OO, Makarova KS, W et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell. 2015 Nov 5;60(3):385-97.[5] Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016 Aug 5;353(6299):aaf5573.[6] East-Seletsky A, O'Connell MR, Knight SC, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016 Oct 13;538(7624):270-273. [7] Smargon AA, Cox DBT, Pyzocha NK, et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol Cell. 2017 Feb 16;65(4):618-630.e7.[8] Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017 Apr 28;356(6336):438-442.[9] East-Seletsky A, O'Connell MR, Burstein D, et al. RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes. Mol Cell. 2017 May 4;66(3):373-383.[10] Chen JS, Ma E, Harrington LB, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018 Apr 27;360(6387):436-439.[11] Gootenberg JS, Abudayyeh OO, Kellner MJ, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018 Apr 27;360(6387):439-444.[12] Myhrvold C, Freije CA, Gootenberg JS, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018 Apr 27;360(6387):444-448.[13] Harrington LB, Burstein D, Chen JS, et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018 Nov 16;362(6416):839-842.[14] Li SY, Cheng QX, Wang JM, et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018 Apr 24;4:20.[15] Li L, Li S, Wu N, et al. HOLMESv2: A CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. ACS Synth Biol. 2019 Oct 18;8(10):2228-2237. [16] Zhou W, Hu L, Ying L, et al. A CRISPR-Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat Commun. 2018 Nov 27;9(1):5012.[17] Pardee K, Green AA, Takahashi MK, et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell. 2016 May 19;165(5):1255-1266.[18] Huang M, Zhou X, Wang H, et al. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Triggered Isothermal Amplification for Site-Specific Nucleic Acid Detection. Anal Chem. 2018 Feb 6;90(3):2193-2200.[19] Zhang Y, Qian L, Wei W, et al. Paired Design of dCas9 as a Systematic Platform for the Detection of Featured Nucleic Acid Sequences in Pathogenic Strains. ACS Synth Biol. 2017 Feb 17;6(2):211-216.[20] Qiu XY, Zhu LY, Zhu CS, et al. Highly Effective and Low-Cost MicroRNA Detection with CRISPR-Cas9. ACS Synth Biol. 2018 Mar 16;7(3):807-813.[21] Li Y, Li S, Wang J, et al. CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol. 2019 Jul;37(7):730-743.
原文地址:https://zhuanlan.zhihu.com/p/445783849 |
|
 /3
/3 